Introduction
Emerging evidence has shed light on the prevalence of germline variants (GV) that increase the risk of myeloid neoplasms in adults. Despite the use of next-generation sequencing (NGS) in the diagnostic workup of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS), establishing a standard-of-care approach to screening patients for GV has been difficult due to a rapidly evolving list of candidate variants and inconsistent referral patterns for confirmatory testing. Here we describe our efforts to characterize molecular features at diagnosis and in remission based on tumor-only sequencing for patients with possible germline predisposition syndromes to myeloid neoplasms.
Methods
We retrospectively analyzed molecular pathology records of adult patients treated at the University of Pennsylvania who were diagnosed with AML and/or MDS between 2013 to 2022, had somatic NGS performed with diagnostic bone marrow biopsy, and had identified variants in genes affiliated with known or emerging MDS/AML germline predisposition syndromes. These variants (Fig. 1A) were based on those defined in ELN 2022 as well curated from the hereditary hematologic malignancy panel at the Children's Hospital of Philadelphia . Analysis focused on pathogenic and likely pathogenic variants.
Results
We reviewed records from 364 patients with a diagnosis of AML (n=311), MDS or MDS/MPN overlap (n=48), or other myeloid neoplasm (n=5). Median age of diagnosis was 66 years (range 21-92 years) with 11% <40 years and 15% <50 years; 56% were male, and 44% were female. Data on race and ethnicity were collected for 331 patients, which reflected demographics typically seen at our center: 86% White, 13% Black, 1% Asian/Pacific Islander, and 4% identified as Hispanic/Latino. Family history data were collected for 316 patients: 13% with hematologic malignancy and/or unexplained cytopenias, 50% with solid tumors, and 2% with a previously known germline predisposition syndrome. 31% of patients had a prior personal history of a non-myeloid cancer.
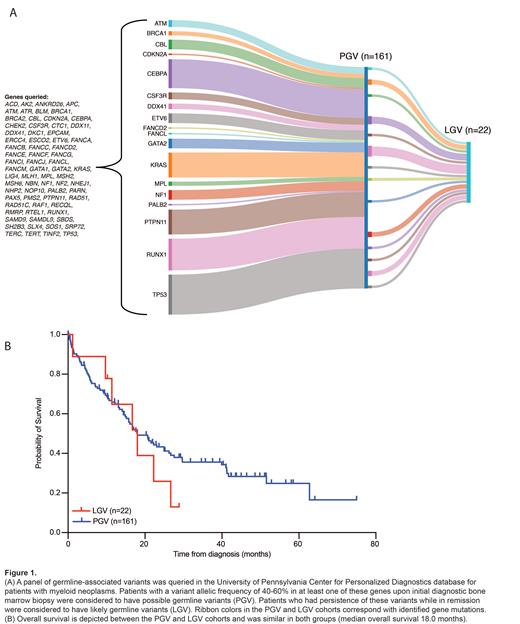
Possible germline-associated variants in our cohort were most frequently seen in TP53 (n=93), RUNX1 (n=49), KRAS (n=48), and PTPN11 (n=47). Only 5 patients had undergone confirmatory exome sequencing via fibroblast culture (2 DDX41, 1 GATA2, 1 TP53, 1 CBL), so we considered patients with a baseline variant allele frequency (VAF) of 30-60% (n=161) on diagnostic studies to have possible GV (PGV) (Fig. 1A). Of these patients, 125 received frontline treatment with intensive chemotherapy (49%), HMA/venetoclax (29%), or HMA or other low-intensity therapy (23%). NGS was performed in remission for 53 patients, of whom 22 (42%) patients had persistence of their previously identified variant, which we classified as likely GV (LGV); 20/22 patients with LGV had persistence of germline-associated VAF >40% in remission. Approximately 6% of patients were <40 years of age in the PGV and LGV groups.
The most frequent LGV were DDX41 (18%), CEBPA (14%), NF1 (9%), BRCA1 (9%), and RUNX1 (9%). Within the LGV cohort, 31% of patients had complex cytogenetics; 61% of patients were classified with ELN 2022 adverse risk or IPSS-R high/very high-risk disease; 64% of LGV patients had a family or personal history of cancer or unexplained cytopenias. Median OS was 18 months for both the PGV and LGV groups (Fig. 1B). When examining co-occurring somatic mutational patterns in the LGV group, we most frequently observed alterations in canonical AML/MDS-associated genes, including ASXL1, FLT3, IDH1/2, NPM1, TET2, and splicing factors.
Conclusions
As the acknowledgement of germline predisposition syndromes increases, it is imperative that suspected GV are recognized by laboratories and clinicians so patients can appropriately be referred for genetic counselling and confirmatory testing. Although confirmatory sequencing in unaffected tissue is considered the gold-standard approach for diagnosis, use of other factors like VAF and persistence of mutational burden in remission can help identify patients with LGV. Indeed, in our cohort, 6% of patients were considered to have LGV, of which 64% had a family or personal history of cancer or unexplained cytopenias. Limitations include the evolution of our somatic sequencing panel throughout the time course evaluated, inability to exclude somatic reversion due to somatic mutational screening only, and exclusion of variants of uncertain significance.
Disclosures
Bhansali:Alva10: Consultancy. Carroll:Cartography Bioscences: Membership on an entity's Board of Directors or advisory committees; Janssen Pharmaceuticals: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal